CRISPR gene-editing technology is a molecular biological process by which the genome of an organism can be cut or edited. It can be also used to modify genetic systems and alter DNA sequences. CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, which are a simplified version of a bacterial defense mechanism that forms the basis for CRISPR-Cas9 genome editing technology. The Cas9 enzyme and synthetic guide RNA complex is delivered into a target cell, allowing the cell genome to be cut or modified at the desired location thus helping in modifying the genome in an organism.
CRISPR technology was discovered by Dr. Jennifer Doudna and Dr. Emmanuel Charpentier. Their paper uncovers that the CRISPR-Cas9 bacterial insusceptible framework which could be repurposed as a gene-editing tool. Though CRISPR was discovered as a gene-editing tech in
2012 still the history of this technology goes back to 1993. It is when Francisco Mojica, a scientist at the University of Alicante in Spain discovered CRISPR in archaea (and later in bacteria). He proposed that CRISPRs serve as part of the bacterial immune system which helps in defending against invading viruses. In January 2013, several papers were published demonstrating how the CRISPR/Cas9 system could be used for gene editing in human cells. To make the CRISPR- Cas9 technique more accurate and efficient changes are currently underway. In 2019, the primary CRISPR clinical preliminaries started by a technique called cell therapy, where the cells from sickle cell diseases (SCD) patients were taken and was in vitro altered prior to instilling them back into the body of the patients. Later, in 2020 for the first time, the promising accomplishment of SCD cell treatment clinical trials, a CRISPR treatment was infused straightforwardly into human patients. This technique is coined as gene therapy, which was utilized for treating inherited blindness. And recently in 2021, a small clinical trial of intravenous CRISPR gene editing was performed in humans which ended with a promising result.
CRISPR technology has a high level of accuracy and is very convenient. They are made of two forms which are based on their specificities like PAM and target sequence. The crRNA array as a part of the CRISPR locus consists of twenty base pairs that act as a target sequence. The base pairs sequence on the host DNA helps the Cas9 proteins to find the accurate location on the host’s genome. The sequence mentioned is not a part of the Cas9 protein thus it can be synthesized and customized independently. PAM sequence cannot be recognized unless the Cas9 is highly modified. Though it’s not a drawback as the PAM sequence is generally a short and nonspecific sequence that occurs throughout the genome. Properly spaced single-stranded
host DNA after single breaking can trigger the homology-directed repair, more accurate than the non-homologous end joining (NHEJ) which is caused by a double-stranded break. The main objective of the HDR process is to utilize the repair template that is available and incorporate
the new sequence that is obtained into the genome. Once administered, the new sequence that has formed has now become a part of the cell’s genetic material which gets passed onto the daughter cells of the successive generation.
How does CRISPR gene editing work?
The CRISPR system includes DNA repairing. While crafted by a microorganism’s protection framework is done once it cuts unfamiliar DNA, genome-altering tool should assemble the pieces of back once more. Cells themselves can perform repairing machinery and join the double stranded breaks in DNA. When a cell gets access to a genetic sequence that is ‘homologous’ to the broken DNA (eg., RNA guide carried by a Cas9 protein) then, at that point, the cell involves that matching nucleotide as a template strand and fills the lacking part by a cycle called Homology-Directed Repair (HDR). In second case, if template is unavailable (as Cas9 tool was designed to cut) then cells perform ligation themselves by joining the cut ends by a process known as ‘Non-Homologous End Joining’ (NHEJ).
Mechanism –
CRISPR-Cas system is an adaptive immune response based on archaea and bacteria. It helps in preventing bacterial infection, conjugation by breaking nucleic acids that enter the cell from outside. A CRISPR was tentatively presented as a critical component in the immune mechanism of prokaryotes against infections. During the interaction, bacterial cells become inoculated by the inclusion of short parts of viral DNA (spacers) into a genomic district called the CRISPR array. Subsequently, spacers act as a hereditary memory of past viral infections. The CRISPR mechanism shields microbes from rehashed viral attack by means of three essential stages: Transformation or Adaptation (spacer procurement), crRNA synthesis (expression), and target obstruction. CRISPR loci are a variety of short tandem repeats and can be seen in chromosomal or plasmid DNA of prokaryotes. Cas is normally viewed as adjoining CRISPR that codes for nuclease protein (Cas protein) capable to obliterate or sever viral nucleic acid.
The CRISPR/Cas-9 genome altering can be classified into three stages: recognition, cleavage, and repair. The sgRNA coordinates with Cas-9 and helps in recognising the target sequence in the gene of interest through its 5ʹ-crRNA base pair. The Cas-9 protein is non-activated without any sgRNA. The Cas-9 nuclease cuts the double stranded DNA at a site 3 base pair upstream to PAM. PAM succession is a short (2-5 base-pair length) conserved sequence present downstream to the cut site and its size fluctuates relying upon the bacterial species. Nuclease is frequently used in the genome-altering apparatus, Cas-9 protein perceives the PAM at 5ʹ-NGG-3ʹ (N can be any nucleotide base). When Cas-9 has found the designated target site, it triggers the surrounding DNA followed by the development of DNA-RNA hybrid. Then the Cas-9 protein initiates the DNA cleavage. HNH area cleaves the base pairing strands, while the RuvC separates the non-base pair strand of target DNA to deliver blunt ended DSBs. At last, the DSB is fixed by the host cell apparatus by repair mechanism.
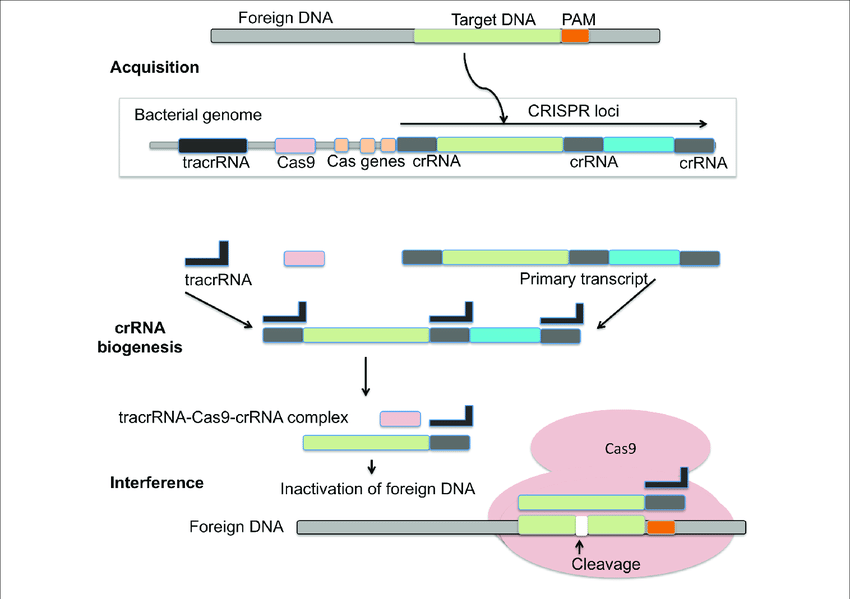
Biogenesis –
To empower insusceptibility, the CRISPR array is transcribed into crRNA (pre-crRNA) that is additionally transformed into mature crRNAs(memorized sequences of invaders). In type I and III frameworks, individuals from the Cas6 family yields crRNAs intermediates that are flanked by a short 5′ tag. One exemption is given by the I-C frameworks, which don’t code for Cas6 proteins. Here, the protein Cas5d processes pre-crRNA bringing about halfway crRNAs with a 11 nucleotide 5′ tag. Further cleavage of the 3′- crRNA intermediates by an obscure nuclease can happen which yields mature crRNA species made out of a full spacer segment (5′ end) and a rehash segment (3′ end), which results into a hair-pin like structure most type I frameworks. The development of crRNAs in class 2 CRISPR-Cas frameworks varies altogether. In type II frameworks, tracrRNA is expected for the handling of the pre-crRNA. The anti-repeat arrangement of this RNA empowers the development of a RNA duplex with pre-crRNA repeats which is balanced out by Cas9. The duplex is then perceived and handled by the host RNase III yielding a crRNA intermediates that goes through additional development leading to the formation of a small guide RNA or sg-RNA. A RNase III-autonomous mechanism was found in II-C type CRISPR-Cas system of Neisseria meningitidis. Here, promoter sequences exists in each repeat and in some case initiated transcription leading to crRNA intermediate species. Despite the fact that RNase III-imediated 3′-crRNA processing: tracrRNA duplex was noticed, it was nonessential for the interference process. In the V-A CRISPR-Cas type, it has been shown that Cpf1 has a double capability during CRISPR-Cas resistance. Cpf1 processes untimely crRNAs followed by a further maturation step that utilizes the crRNAs that it has created to cleave the target DNA sequence.

References –
Redman M, King A, Watson C, King D (August 2016). “What is CRISPR/Cas9?”.
Archives of Disease in Childhood: Education and Practice Edition. 101 (4): 213–215. doi:10.1136/archdischild-2016-310459 PMC 4975809. PMID 27059283
Zhang F, Wen Y, Guo X (2014). “CRISPR/Cas9 for genome editing: progress, implications and challenges”. Human Molecular Genetics. 23 (R1): R40–6. doi:10.1093/hmg/ddu125 PMID 24651067
CRISPR-CAS9, TALENS and ZFNS – the battle in gene editing https://www.ptglab.com/news/blog/crispr-cas9-talens-and-zfns-the-battle-in- gene-editing/
Kaiser J (26 June 2021). “CRISPR injected into the blood treats a genetic disease for first time”. Science | AAAS. Retrieved 11 July 2021.
Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, et al. (August 2021). “CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis”. The New England Journal of Medicine. 385 (6): 493–502. doi:10.1056/NEJMoa2107454. PMID 34215024. S2CID 235722446.
Boonstra E, de Kleijn R, Colzato LS, Alkemade A, Forstmann BU, Nieuwenhuis S (6 October 2015). “Neurotransmitters as food supplements: the effects of GABA on brain and behavior”. Frontiers in Psychology. 6: 1520. doi:10.3389/fpsyg.2015.01520. PMC 4594160. PMID 26500584.
“Tomato In Japan Is First CRISPR-Edited Food In The World To Go On Sale”. IFLScience. Retrieved 18 October 2021.
Hsu PD, Lander ES, Zhang F (June 2014). “Development and applications of CRISPR-Cas9 for genome engineering”. Cell. 157 (6): 1262–1278. doi:10.1016/j.cell.2014.05.010. PMC 4343198. PMID 24906146.“Press release: The Nobel Prize in Chemistry 2020”. Nobel Foundation.
Wu KJ, Peltier E (7 October 2020). “Nobel Prize in Chemistry Awarded to 2 Scientists for Work on Genome Editing – Emmanuelle Charpentier and Jennifer A. Doudna developed the Crispr tool, which can alter the DNA of animals, plants and microorganisms with high precision”. The New York Times.
Kunin V, Sorek R, Hugenholtz P (2007). “Evolutionary conservation of sequence and secondary structures in CRISPR repeats”. Genome Biology. 8 (4): R61. doi:10.1186/gb-2007-8-4-r61. PMC 1896005. PMID 17442114
Wright AV, Nuñez JK, Doudna JA (January 2016). “Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering”. Cell. 164 (1–2): 29–44. doi:10.1016/j.cell.2015.12.035. PMID 26771484
Jensen TI, Axelgaard E, Bak RO (June 2019). “Therapeutic gene editing in haematological disorders with CRISPR/Cas9”. British Journal of Haematology. 185 (5): 821–835. doi:10.1111/bjh.15851. PMID 30864164. S2CID 76663873.
Reardon S (2016). “First CRISPR clinical trial gets green light from US panel”. Nature. doi:10.1038/nature.2016.20137. S2CID 89466280
Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF (February 2012). “Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity”. Molecular Cell. 45 (3): 303–313. doi:10.1016/j.molcel.2011.12.013. PMC 3381847. PMID 22227115.
Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (November 2009). “RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex”. Cell. 139 (5): 945–956. doi:10.1016/j.cell.2009.07.040. PMC 2951265. PMID 19945378
Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, et al. (February 2008). “Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus”. Journal of Bacteriology. 190 (4): 1401–1412. doi:10.1128/JB.01415-07. PMC 2238196. PMID 18065539.
Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S (February 2008). “Phage response to CRISPR-encoded resistance in Streptococcus thermophilus”. Journal of Bacteriology. 190 (4):1390–1400. doi:10.1128/JB.01412-07. PMC 2238228
Shah SA, Erdmann S, Mojica FJ, Garrett RA (May 2013). “Protospacer recognition motifs: mixed identities and functional diversity”. RNA Biology. 10 (5): 891–899.
Doudna JA, Charpentier E (November 2014). “Genome editing. The new frontier of genome engineering with CRISPR-Cas9”. Science. 346 (6213): 1258096. doi:10.1126/science.1258096. PMID 25430774. S2CID 6299381.
Ledford H (June 2015). “CRISPR, the disruptor”. Nature. 522 (7554): 20–24.
Bibcode:2015Natur.522…20L. doi:10.1038/522020a. PMID 26040877.
Hsu PD, Lander ES, Zhang F (June 2014). “Development and applications of CRISPR-Cas9 for genome engineering”. Cell. 157 (6): 1262–1278. doi:10.1016/j.cell.2014.05.010. PMC 4343198. PMID 2490614
Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150496. doi: 10.1098/rstb.2015.0496. PMID: 27672148; PMCID: PMC5052741.
Souhrid Sarkar
Souhrid Sarkar is an Aspiring Biotechnologist. He is a B.Tech student at Amity University Kolkata. He is content writer at BioXone. He is a Research Intern at Indian Institute of Technology, Bombay (IITB) & Reviewer at TMR Publishing Group. He is Keyboardist by hobby.

Pingback: CRISPR - Methods, Techniques & Applications - BiokiMicroki