Bacterial DNA replication is an essential step for reproduction – Binary fission.
Introduction:
DNA replication is a process in which organisms duplicate their DNA molecule or make its exact copy in order to transfer the genetic information into its daughter cell (next generation). Hence, DNA replication occurs before cell division. DNA replication ensures that both parental and daughter cell have the exact copy of its genetic material.
To revise the DNA replication process we suggest you to read our article DNA Replication Process – A quick Read.
The Bacterial DNA replication of bacteria is well understood than eukaryotes because of the following reasons-
- The bacterial genome is small as compared to eukaryotes
- The bacterial chromosome is less condensed than eukaryotes.
- The DNA replication of eukaryotes is complex as compared to bacterial DNA replication.
- The rate of bacterial DNA replication is very fast as compared to eukaryotic DNA replication.
To understand bacterial DNA replication,
Characteristics of E. Coli Chromosome:
- It has single chromosome
- It has Covalently closed circular chromosome (CCC)
- The size of E.coli genome is 4.6Mbp (Million base pairs).
- It has poly and monocistronic genes.
DNA Replication of E. coli
Prokaryotic DNA replication is well studied in the model microorganism – E. coli. The DNA replication process is divided into three steps namely –
- Initiation
- Elongation
- Termination
These steps are divided and can be differentiated on the basis of the reactions occurring and the enzymes involved in different steps.
Initiation of DNA replication in E. coli
- It means beginning of DNA replication.
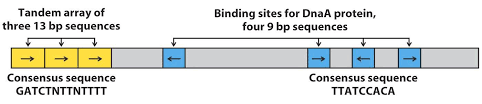
- The site where replication begins is called as origin of replication ‘ori C’ (C stands for coli, it is named because it is isolated from E. coli).
- The oriC consists of 245 bp and it has conserved sequence, found in bacterial chromosome
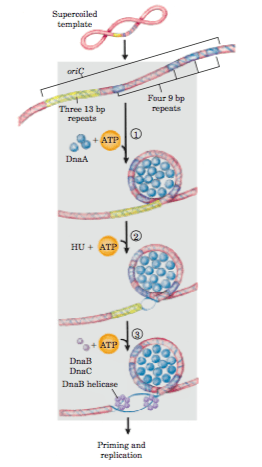
- There are two key series of short sequences present in oriC that dictates DNA replication initiation. The first one is 3 repeats of 13 bp long sequence ((It is AT rich region) and second is 4 repeats of 9 bp long sequence.
- The four to five molecules of DNA replication initiator protein Dna A binds to four repeats of 9bp. Thus it can be said that the four repeats of 9 bp is site of recognition for Dna A protein.
- The binding of dna A protein to 9 bp repeats causes denaturation at the three repeats of 13 bp forming replication bubble. Denaturation requires energy from ATP and assistance of HU(histone like proteins).
- The denaturation causes formation of replication bubble and thus forming two replication forks, which will move in clockwise and anticlockwise direction respectively.
- In the next step, Dna c protein assists in loading Dna B (helicase) at the unwound region.
- On each strand of DNA a ring like hexamer of Dna B, unwinds the DNA bi-directionally.
- SSB proteins stabilize the exposed single strands of DNA.
- The topological strain caused by unwinding of DNA is relieved by DNA gyrase (DNA TOP II).
- It is highly regulated process. It occurs only once during each cell division. The GATC sequence present 11 times in oriC. The adenine of GATC is methylated by Dam methylase (DNA adenine methylation). When only strand is methylated it is called as hemimethylated and when both the strands are methylated called as fully methylated. The fully methylated state serves as an active origin. Hence, the process is initiated only when GATC is fully methylated. Slow hydrolysis of ATP by Dna A protein also regulates the replication.
Elongation of DNA Replication
- It means extension of DNA strands.
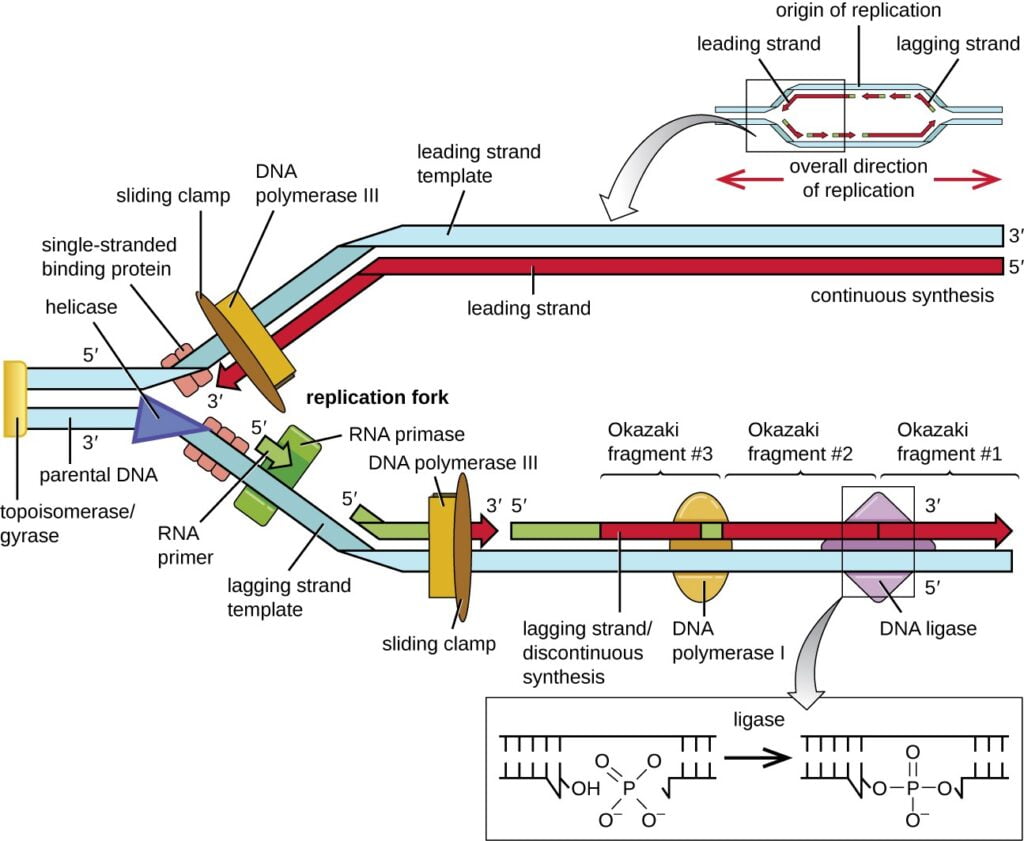
- It is done by addition of nucleotides at 3′ end of growing strand by DNA Polymerase III.
- One strand is continuously synthesized in 5′-3′ direction, so it is called leading strand.
- The other strand is dis continuously synthesized in 5′-3′ direction, and rate of synthesis is slow hence it is called lagging strand.
- As DNA polymerase can only elongates the DNA strand but cannot initiate. Hence, DNA polymerase is need a primer for DNA polymerization. Dna G protein acts as primase and add RNA primer at site of origin.
- DNA Polymerase III add the new Deoxyribonucleotides continuously on leading strand. The complex of Dna B and Dna G is called as primosome.
- The single DNA polymerase can synthesize both DNA strands. It is possible because of its asymmetric structure. The DNA polymerase enzyme has two core subunits. One subunit elongates leading strand while other elongates lagging strand.
- On lagging strand, primase gets associated occasionally with DNA helicase for synthesizing RNA primer. The clamp-loading complex of DNA polymerase III positions the beta sliding clamp at the primer forming clamp loading complex. A short DNA strand is synthesized called as okazaki fragment. After every formation of an okazaki fragment, DNA polymerase dissociate from former beta sliding clamp and associates with new beta sliding clamp, this allows synthesis of new okazaki fragment.
- Once the synthesis of okazaki fragment is complete, DNA polymerase I removes the primer and replace it with complementary deoxyribose nucleotides called as Nick translation. The Ligase enzyme reseals the gap.
- All the coordinated components involved in DNA synthesis at the replication site forms a complex and called as Replisome.
- After completion, the primers are removed by Dna Polymerase I and the Nick’s are sealed by Dna ligase.
Termination
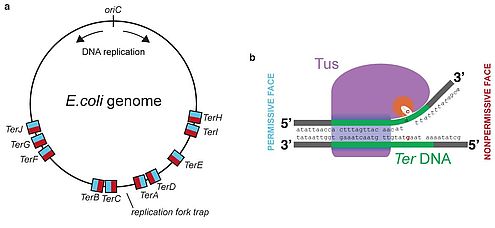
- It means the end of replication process.
- It occurs in the region called Ter region and it is about 20bp.
- The Tus proteins bind with the Ter region and form Ter-Tus complex.
- The Ter-Tus complex works as a trap in unidirection and halts one of the replication fork.
- The other replication fork gets halted on colliding with the previously halted fork.
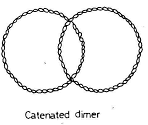
- The newly formed circular DNA remains interlinked to each other and it is called as catenanes.
- These covalently closed circular chromosomes are separated by Topoisomerase IV and passed on to daughter cell.
Activity:
After reading the article, write the function of enlisted proteins of bacterial DNA replication.
| Dna A protein | |
| Dna B protein (Helicase) | |
| Dna C protein | |
| HU (Histone like protein) | |
| Dna G protein (primase) | |
| Single stranded binding protein | |
| DNA gyrase | |
| Dam methylase |
References:
http://utminers.utep.edu/rwebb/html/the_e__coli_genome.html
https://www.wikigenes.org/e/gene/e/960427.html
Principle of Biochemistry, Nelson and Cox, 5th Edition
B. DNA Replication in Bacteriafaculty.ccbcmd.edu › nucleoid › nucleoid › nucleoid3
very helpful , its gives the whole idea of how DNA replication occurs in prokaryotes.
Pingback: Molecular Animation: Future of teaching Biology - BiokiMicroki