INTRODUCTION
CRISPR is a specific, efficient and versatile gene-editing technology we can harness to modify, delete or correct precise regions of our DNA.
Dr. Emmanuelle Charpentier & Dr. Jennifer Doudna co-invented CRISPR/Cas9 gene editing.
In the year 2020 both the scientists who pioneered the revolutionary gene-editing technology got Nobel Prize in Chemistry. Until then, people knew “CRISPR” only as an acronym for the Clustered Regularly Interspaced Short Palindromic Repeats of genetic information that some bacterial species use as part of an antiviral mechanism. Now, as a gene-editing tool, CRISPR/Cas9 has revolutionized biomedical research and may soon enable medical breakthroughs in a way few biological innovations have before. CRISPR/Cas9 edits genes by precisely cutting DNA and then letting natural DNA repair processes to take over. The system consists of two parts: the Cas9 enzyme and a guide RNA.
CRISPR Lexicon :
- CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats of genetic information that some bacterial species use as part of an antiviral system. Scientists discovered how to use this system as a gene-editing tool.
- Cas9: a CRISPR-associated (Cas) endonuclease, or enzyme, that acts as “molecular scissors” to cut DNA at a location specified by a guide RNA.
- Deoxyribonucleic acid (DNA): the molecule that most organisms use to store genetic information, which contains the “instructions for life”.
- Ribonucleic acid (RNA): a molecule related to DNA that living things use for a number of purposes, including transporting and reading the DNA “instructions”.
- Guide RNA (gRNA): a type of RNA molecule that binds to Cas9 and specifies, based on the sequence of the gRNA, the location at which Cas9 will cut DNA.
- protospacer adjacent motif (PAM): PAM is an essential targeting component which distinguishes bacterial self from non-self DNA. No editing can occur at any site other than one at which Cas9 recognizes PAM.
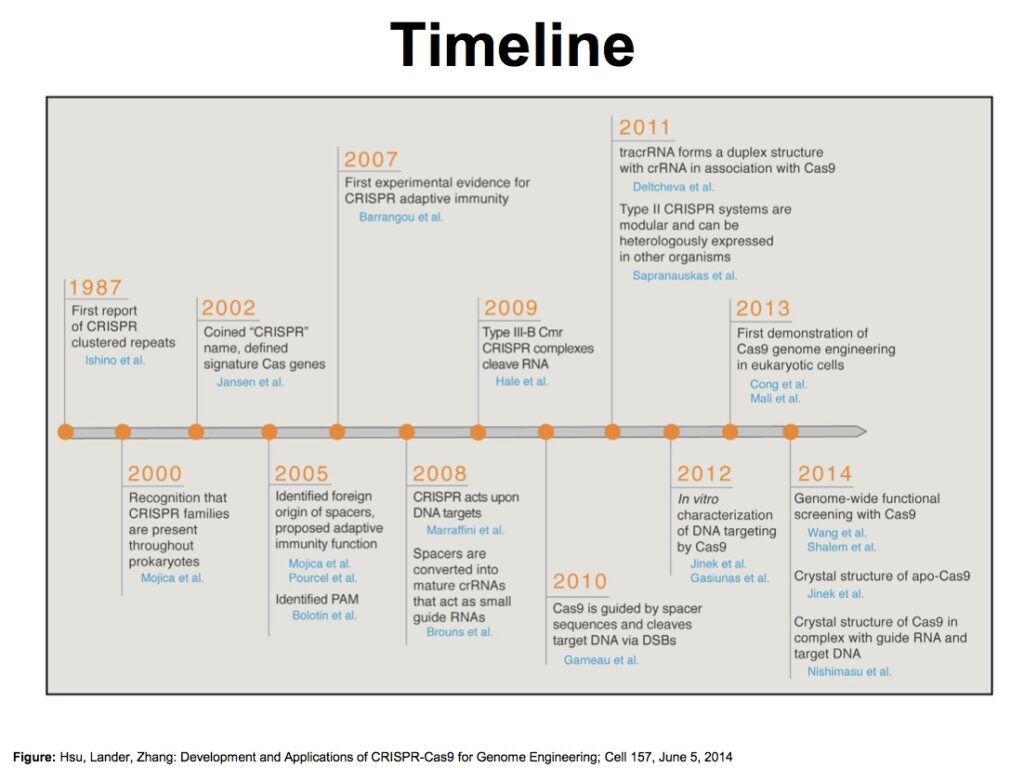
How does it work ?
- The system consists of two parts: the Cas9 enzyme and a guide RNA.
- Cas9 acts as a pair of ‘molecular scissors’ that can cut the two strands of DNA at a specific location in the genome so that bits of DNA can then be added or removed.
- gRNA consists of a small piece of pre-designed RNA sequence (about 20 bases long) located within a longer RNA scaffold. The scaffold part binds to DNA and the pre-designed sequence ‘guides’ Cas9 to the right part of the genome. This makes sure that the Cas9 enzyme cuts at the right point in the genome.
- The guide RNA has RNA bases that are complementary to those of the target DNA sequence in the genome. This means that the guide RNA will only bind to the target sequence and no other regions of the genome.
- The Cas9 follows the guide RNA to the same location in the DNA sequence and makes a cut across both strands of the DNA.
- At this stage the cell recognises that the DNA is damaged and tries to repair it.
- Scientists can use the DNA repair machinery to introduce changes to one or more genes in the genome of a cell of interest.[fig.2]
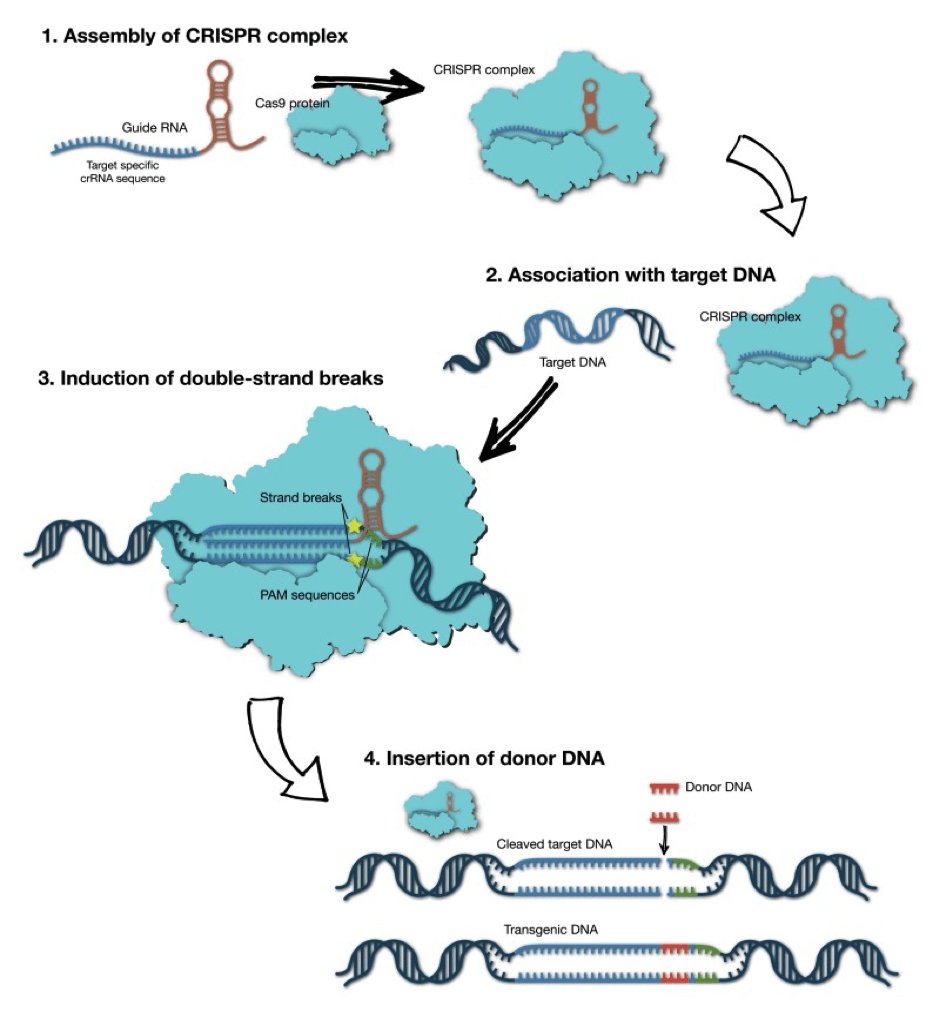
Three main categories of genetic edits can be performed with CRISPR/Cas9:
- DISRUPT :- If a single cut is made, a process called non-homologous end joining can result in the addition or deletion of base pairs, disrupting the original DNA sequence and causing gene inactivation.
- DELETE :- A larger fragment of DNA can be deleted by using two guide RNAs that target separate sites. After cleavage at each site, non-homologous end joining unites the separate ends, deleting the intervening sequence[fig.3]
- CORRECT OR INSERT:- Adding a DNA template alongside the CRISPR/Cas9 machinery allows the cell to correct a gene, or even insert a new gene, using a process called homology directed repair. [fig.3]

APPLICATION OF TECHNIQUE:-
- Gene silencing[9,10]
- DNA-free CRISPR-Cas9 gene editing
- Homology-directed repair (HDR)[7,8]
- Transient gene silencing or transcriptional repression (CRISPRi)[11,12]
- Transient activation of endogenous genes (CRISPRa or CRISPRon)[12,13]
- Embryonic stem cell and transgenic[14,15]
- Pooled genome-scale knockout screening
REFERENCES
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D.A. Romero, P. Horvath CRISPR provides acquired resistance against viruses in prokaryotes Science, 315 (2007), pp. 1709-1712
- Y. Ishino, H. Shinagawa, K. Makino, M. Amemura, A. Nakata Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product J. Bacteriol., 169 (1987), pp. 5429-5433
- R. Jansen, J.D. Embden, W. Gaastra, L.M. Schouls Identification of genes that are associated with DNA repeats in prokaryotes Mol. Microbiol., 43 (2002), pp. 1565-157
- F.J. Mojica, C. Díez-Villaseñor, E. Soria, G. Juez Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria Mol. Microbiol., 36 (2000), pp. 244-246
- C. Pourcel, G. Salvignol, G. Vergnaud CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies Microbiology, 151 (2005), pp. 653-663
- H. Wang, H. Yang, C.S. Shivalila, M.M. Dawlaty, A.W. Cheng, F. Zhang, R. Jaenisch One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineerin Cell, 153 (2013), pp. 910-918
- Multiplex genome engineering using CRISPR/Cas systems. Science, 2013. 339(6121): p. 819-23
- Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 2013. 154(6): p. 1380-9.
- RNA-programmed genome editing in human cells. Elife, 2013. 2: p. e00471.
- RNA-guided human genome engineering via Cas9. Science, 2013. 339(6121): p. 823-6.
- Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 2013. 152(5): p. 1173-83.
- CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 2013. 154(2): p. 442-51.
- Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res, 2013. 23(10): p. 1163-71.
- One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 2013. 153(4): p. 910-8.
- Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell, 2014. 156(4): p. 836-43.
- Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014. 343(6166): p. 80-4.
- Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 2014. 343(6166): p. 84-7.


One thought on “Sharpest Tool of Genetic Engineering: CRISPR-Cas9”