Introduction –
Transcription is the synthesis of RNA from DNA, catalysed by a very special enzyme, RNA polymerase (RNAP). The DNA in the gene is used as a template to make a Messenger RNA (mRNA) strand which is done with the help of RNAP.
RNAP was Discovered by Samuel B. Weiss and Jerard Hurwitz in 1960. It is also known as DNA dependent RNA polymerase. It binds to the promoter sequence and moves from the promoter region to the terminator region. Upon binding to the promoter sequence, it creates a Transcription Bubble, which then moves along the DNA, and the RNA chain keeps growing in length. Within this bubble, RNAP catalyse and scrutinise the process of addition and pairing of base pairs.
RNAP in Prokaryotes and Eukaryotes –
RNA polymerase in prokaryotes
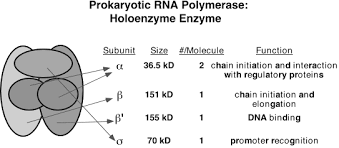
In Prokaryotes, only one type of RNAP is involved in the synthesis of different types of RNA (rRNA, mRNA, tRNA, etc). RNAP, also referred to as RNA Polymerase Holoenzyme, is a multicomplex enzyme with a molecular weight of 465kD. The polymerisation rate of RNAP in majority of the prokaryotes is 40-50 nucleotide per second.
Being a multicomplex enzyme, it’s made up of five different subunits (polypeptides).
The RNAP holoenzyme is divided into two components, namely the core enzyme and the sigma factor.
The core enzyme includes two same α, β, β’ and ω subunit.
The sigma factor includes σ subunits.
RNAP holoenzyme can be inhibited by an antibiotic called RIFAMPICIN.
Now let us look at all the subunits individually on what roles they play and what are the genes responsible for encoding them.
α subunit –
- It is encoded by a gene called rpoA gene.
- They are in pair i.e., two alpha subunits.
- It is located in core enzyme.
- Its function is to bind to the promoter region and involved in regulation of transcription.
- Molecular weight is about 41Kd.
β subunit –
- They are encoded by rpo B genes.
- Molecular weight is 155kd.
- Located in core enzyme.
- Functions includes as catalytic site and binds to NTPs (nucleotide tri-phosphates), such as ATP, UTP, GTP etc.
- Inhibited by Rifampicin, an Antibiotic which inhibits the holoenzyme not the core enzyme.
β’ subunit –
- They are encoded by rpo C gene and have molecular weight of 165 Kd.
- Located in the core enzyme.
- It facilitates the catalytic activity after binding to the DNA template strand.
- Generally, it binds to Zn+ ions.
ω subunit –
- Encoded by rpo Z gene.
- Located in the core enzyme.
- Functions include assembly of RNA polymerase.
- It is the smallest subunit as its molecular weight is about 10kd.
- It contains 91 Amino acids.
σ subunit –
- This subunit is encoded by rpo D genes (sigma 70).
- located in the holoenzyme.
- Functions include, recognition of promoters and increase the Affinity of holoenzyme to bind to promoters.
The σ70 is known as the “housekeeping” factor, because it initiates transcription of most genes.
Bacterial cells have different types of σ factors which is encoded by different types of genes and also different function.
- σ54 is encoded by rpoN gene and involves in nitrogen regulated gene transfer.
- σ38 is encoded by rpoS gene and involves in gene expression during stationary and starvation phase.
- σ32 is encoded by rpoH gene and is involved in heat shock gene transcription.
RNA polymerase in eukaryotes
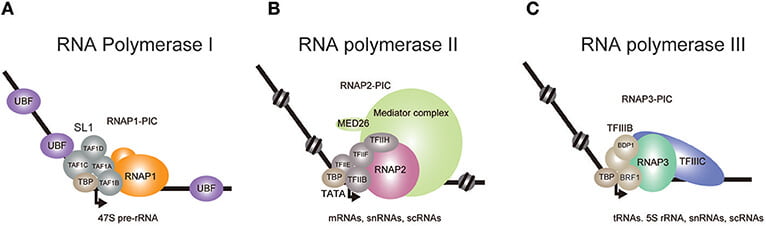
In eukaryotes, there are five different types of RNA polymerase from which three are main, they are RNA polymerase I, RNA polymerase II and RNA polymerase III. Well, a question arises that how are these polymerases identified? and on what basis are they differentiated?
The identification of these RNA polymerases was done using a toxin known as α-amanitin. α-amanitin is extracted i.e. isolated from a poisonous mushroom called Amanita phalloids. It is made up of eight amino acids and is a cyclic peptide. It is also known as Death cap.
- RNA polymerase II and III are inhibited by this toxin, whereas RNA polymerase I is not.
- RNA polymerase II is highly sensitive and is inhibited at concentration of 1μg/ml of α-amanitin.
- RNA polymerase III is moderately sensitive and is inhibited at concentration of 10μg/ml of α-amanitin.
- All the eukaryotic RNA polymerases have molecular mass more than 500kDa.
Now, let us see all the RNA polymerase individually.
- RNA polymerase I – responsible for synthesizing rRNA, which is the pre rRNA 45s which matures into 28s, 18s and 5.8s rRNAs. Then forms the major section of ribosomes.
- RNA polymerase II – responsible for synthesizing precursor of mRNA. Also responsible for some snRNAs and microRNAs. Contains about 12 subunits, where the largest two subunits resemble to the bacterial β and β’ subunits.
- RNA polymerase III – responsible for synthesizing tRNAs, 5srRNAs and other small RNAs.
The other two types of RNA polymerases are less understood and are mainly present in plants. Responsible for synthesizing siRNA.
The production of rRNA in the cell is about 90-95%, whereas mRNA and tRNA is about 2% and 3% respectively.
Transcription factors
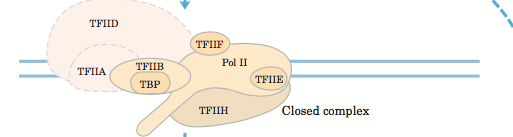
There are various general transcription factors which helps RNA polymerase bind to the promoter region during Transcription similar to the σ factors in bacteria. These factors also help in gene expression. They are proteins in nature.
For instance, TFIID [Transcription factor II D] is responsible for binding to the TATA region present on the promoter sequence. It contains TATA-box binding proteins also known as TBP. These proteins are among the most important transcription factors required to assemble the initiation complex.
There are other factors as well like, TFII [H, E, F, A, B]. why are these factors named A, B, E etc.? well, the answer is that the researcher who discovered this GTFs (general transcription factors) took the Nuclear extract and spilt it into four fractions and these four fractions were named as A, B, C and D. Then each fraction was eluded with a different eluate. So, from fraction A came a protein and named it as TFIIA. similarly, from protein D came TFIID. Likewise, from fraction B, nothing came out but from fraction C, came four different proteins TFIIE, TFIIF, TFIIH and TFIIB. There are also other proteins like TFIIJ and TFIIG which are not always required and are not of great importance.
Another types of transcription factors are also present, which are known as specific transcription factors which binds with the enhancers and increase the transcription rate.
References –
https://pathfinderacademy.in/book/csir-ugc-net-life-sciences-theory-practice-book-19.html
Karp’s Cell and Molecular Biology, 9th Edition.


2 thoughts on “RNA Polymerase in Prokaryotes and Eukaryotes”