Transcription is the first step for protein synthesis. It is the process by which the m-RNA is synthesized and the information is transferred from nucleus to cytoplasm. Followed by Translation synthesizing polypeptide chain.
Introduction –
Transcription is the process were RNA is synthesised from DNA with the help of an enzyme called RNA polymerase (RNAP). Transcription in prokaryotes is quite simple than eukaryotes. Different polymerase enzymes and post transcriptional modification of mRNA is what makes eukaryotic transcription intricate. For better understanding about what RNA polymerase is, do check this Article https://biokimicroki.com/rna-polymerase-in-prokaryotes-and-eukaryotes/.
In transcription, RNA chain (m-RNA, t-RNA, or r-RNA) are synthesized identical to one strand of DNA (the coding strand). The other strand act as a template (also known as non-coding strand or Antisense strand). Further, when the m-RNA (polynucleotide chain) synthesizes protein (polypeptide chain) and this process is called as Translation. The DNA replication, Transcription, Translation are all together called as central Dogma of life.
In this article, We’ll be looking in detail about the transcription process prokaryotes and its inhibitors.
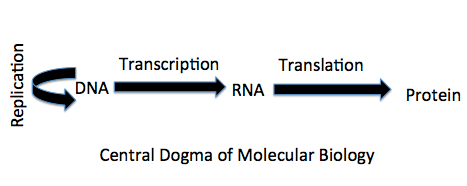
DNA replication, transcription and translation have similar steps namely initiation, elongation and termination but their purpose is different. DNA replication synthesizes DNA, Transcription synthesizes RNA and Translation synthesizes polypeptide chain. All together the process is called as Central Dogma of Life. The Replication is carried out by DNA polymerase and Transcription is processed by RNA polymerase. In DNA replication, multiple types of DNA polymerase are needed whereas for transcription, a single polymerase carries out the process. One more difference between the polymerases is, that DNA polymerase is dependent on primer whereas the RNA polymerase is not.
In English, transcription means a process in which audio or speech is converted into a written document. While translation means conversion of verbal or written text from one language to another. Can you try to relate the usage of these terms in central dogma of life?
The information of Gene from DNA is converted in to RNA form. And the message is written in the same language i.e. language of nucleotide sequence (DNA and RNA are polynucleotide chains). Hence, it is called as Transcription. While, the information in RNA is converted to protein (amino acid chain), changing the language from nucleotide sequence to amino acid sequence.
Let’s discuss the steps and process of transcription.
Transcription involves three steps –
1] Initiation
2] Elongation
3] Termination
Initiation –
Initiation is the first step to start transcribing the mRNA strand and is the most important step. It is also the rate determining step.
Series of steps are involved in this process –
1] Formation of closed complex.
2] Transition of closed complex to open complex.
3] Formation of ternary complex.
Now, before looking into these steps, we need to know the structure of gene
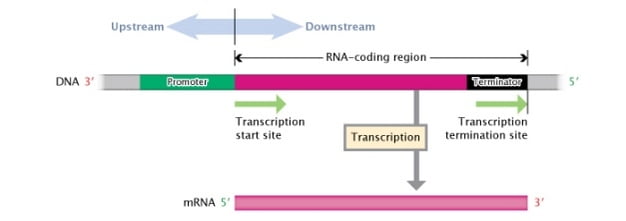
The coding sequence of gene starts with +1 site also known as TSS (transcription start site).
The region or sequence which is present before the TSS is known as upstream region and the sequence which is present after is known as downstream sequence or region.
The sequences present before TSS is known as Promoter.
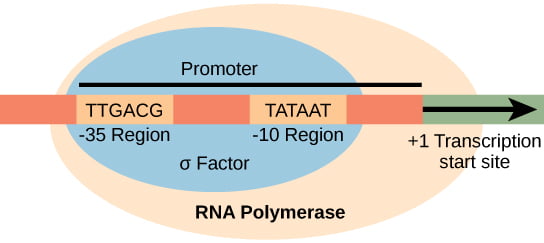
The promoter has -35 region and -10 region present where the sequences in the -35 region is TTGACA and in -10 is TATAAT, which are consensus sequence (i.e conserved sequence which has similar structure and function in different organisms). The sequence located in -10 region is also called as Pribnow box (Pribnow-Schaller box). It is named after David Pribnow and Heinz Schaller.
In some bacteria, there is no presence of -35 region and instead there is an extended -10 region, which have a consensus sequence of TRTG. Some bacteria may also have UPE region also known as upstream element region.
RNA polymerase core enzyme can bind anywhere in the DNA but sigma factors are responsible for the core enzyme to bind to the consensus sequence, at the promoter region. Thus, known as the RNA Polymerase (RNAP) Holoenzyme (core enzyme + sigma factor). Types of sigma factors varies in bacterial species. The sigma factors are distinguished based on their molecular weight. E.coli has 7 different types of sigma factors.
In E. coli, the σ70 is responsible for the recognition of the promoter region. Its molecular weight is 70KDa and hence it is named accordingly.
Sigma factor have four domains σ1, σ2, σ3 and σ4. Each domain recognises a particular region, sigma domain 2 and sigma domain 4 recognises -10 region and -35 region respectively. The distance between both the domains is about 70 Angstrom. The extended -10 region is recognized by sigma domain 3. The upstream element (UPE) region is not recognized by the sigma domain but instead it is recognized by the αCTD (Carboxyl terminal domain) region present in the core enzyme. The other end αNTD (Amino terminal domain) is joined by the linker protein and it is embedded within the core enzyme.
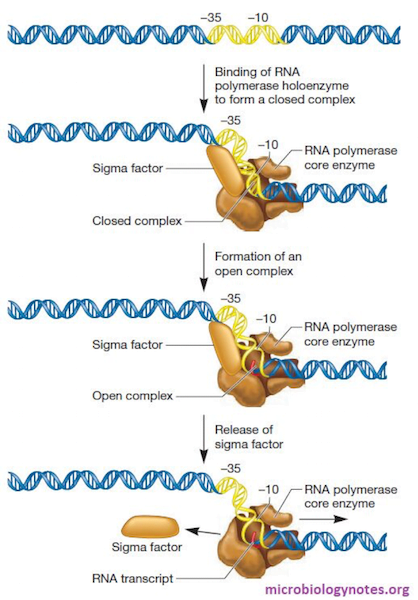
So, the formation of closed complex involves the binding of the holoenzyme to the promoter region.
Generally, the closed complex is reversible i.e. they can dissociate from the promoter region. Once the polymerase binds to the promoter region, the span of the holoenzyme is between [-55 region to +1 site].
Here, melting of DNA or separation of strands does not occur, therefore the DNA remains double-stranded. As, it is reversible it is denoted by equilibrium constant (KB). Since, it is sequence- dependent, there are wide range of values of constant.
In the transition from closed complex to open complex happens unwinding of DNA occurs.
In open complex, the DNA is melted (spanning from -11 region till +3 region). Thus, forming a transcription bubble. The melting occurs at -10 region of the enzyme. The sigma domain 2 (σ2) is responsible for melting. About 1.3 turn of double helix is unwounded. This structural transition is also called isomerization. There is an exception in some promoters (e.g., rRNA promoters) where stable open complexes are not formed.
The open complex is irreversible i.e. it cannot be dissociated from the promoter sequence. Thus, we can denote this reaction with forward rate constant (kf).
Structural changes do occur during this step, about ~90 degrees bending of DNA happens. The sole reason for this structural change is that it allows the template strand to move towards the active site of the holoenzyme. Further, the open complex extends from -55 to +20 region.
Basically, there are five channels in open complex, the RNA exit channel, the ribonucleotide uptake channel, and the other three are involved in the entry and exit of DNA.
Once open complex is formed, the first two nucleotides gets incorporated and phosphodiester bonds are also formed between them.
Thus, formation of ternary complex is formed, as it contains RNA, DNA, and the enzyme. There is a probability that few chains of RNA might release which leads to Abortive initiation.
It’s still not clear that why this abortive initiation takes place but a sigma domain is involved here (σ3.2) which acts as a molecular mimic of RNA which needs several attempts to be removed as it lies in the middle of the RNA exit channel which does not allow the RNA nucleotide chain to grow.
when more than 10 nucleotides get attached, a stable ternary complex is generated and is ready to go for elongating phase.
Elongation –
We have seen five channels in the open complex, where the DNA enters into the catalytic site i.e. the β and β’, from one end and exits from the other end forming a bubble. The RNA polymerase read the DNA template and add RNA nucleotides forming a polynucleotide chain. This process continued until it reaches stop point.
Various proteins are also involved in this process like the rudder proteins, lid proteins, zipper proteins in the exit channel.
In Eukaryotes, the proof reading is carried by TFIIS factor of RNA polymerase II but in porkaryotes, it is done by pyrophosphorolytic editing and Hydrolytic editing. The hydrolytic editing activity is carried out by Gre factors, which are also involved in the stimulation of elongation phase.
Other proteins like the Nus proteins are involved in the regulation of transcription as well.
Termination –
There are sequences in DNA which ends the process of transcription and it is called as termination.
In prokaryotes, two types of termination have been observed:
- Intrinsic termination
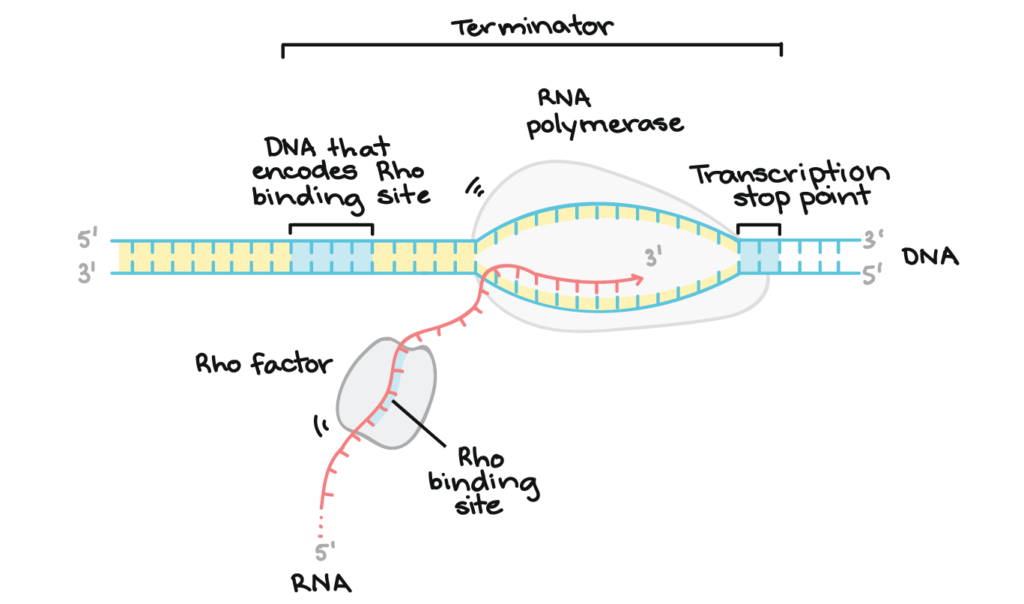
- Rho-dependent termination
Intrinsic termination also known as Rho independent termination which does not require any Rho proteins. Specific sequences are involved which eventually terminate the RNA transcripts. Formation of hairpin loop is observed in this type of termination. The formation of hairpin loop is due to G-C rich region as they have a high affinity to form bonds. This G-C rich sequence is followed by the A-U stretches, as A-U are the weakest of all the base pair thus termination occurs as they are disrupted by the hairpin loop.
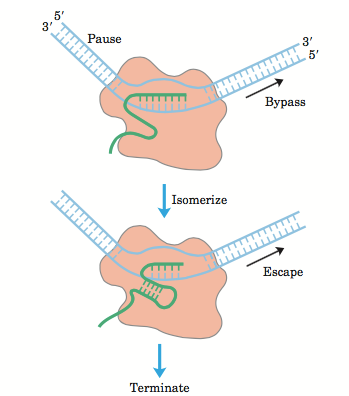
Rho-dependent termination involves the Rho protein which is a ring like structure, which consists of six identical subunits. There are ‘Rut’ sites where it binds to the transcript and, with the ATP driven energy it moves upwards and cleave off the RNA transcript. The Rut sites are rich in C residues and stretch about 40 nucleotides. Secondary structure formation does not happen as they remain largely single stranded.
Inhibitors of Transcription –
Rifampicin – An antibiotic drug which inhibits the prokaryotic transcription. They do not inhibit eukaryotic transcription. They generally bind to the beta subunit of the polymerase. used in the treatment of leprosy and tuberculosis.
Actinomycin-D – They inhibit both DNA synthesis and RNA synthesis by blocking the elongation chain. They are used as anti-cancer drugs.
References –
- Lewin’s genes XI… [Jocelyn E Krebs; Benjamin Lewin; Stephen T Kilpatrick; Elliott S Goldstein] .
- “Molecular Biology of the Gene,” James Watson et. al, 5th Edition.


Heartily thanks for such amazing information