For each chromosome contributed by the sperm there is corresponding chromosome contributed by egg, there was two chromosome of each kind, which together constitute a pair – Thomas Hunt Morgan
While teaching, I have observed that students always get confused between the term and meaning of DNA, gene, chromatin, chromosome and relevant terms. Reading this article, will help the reader to overcome this confusion. Let us understand the term individually. The article also elaborate about the packaging of DNA.
Fundamantals of Chromosome –
DNA –
It is a double helical structure made up of two anti-parallel polynucleotide strands.
Gene –
It is a segment of DNA that contains information for synthesizing proteins or RNA. Hence, it is part of DNA that is carrying the vital information.
Chromosome –
The chromosome is the most compact and condensed form of DNA. The chromosome form of DNA is required for its equal partition into daughter cells. This condensation is accomplished by folding the chromatin fibers into loops, coils and supercoils. To understand the extent of packaging and coiling, imagine a rope of length equal to the field length of the football playing ground and this rope is coiled and condensed to half an inch ball. What a beauty!!!
Junk DNA – The segments or parts of DNA which do not carry any information of synthesizing protein or RNA is called junk DNA. It is non coding region. Such junk DNA are interspersed between the genes. It acts like spaces and its true function is yet to be discovered clearly.
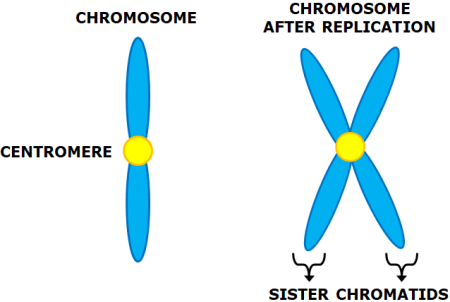
Sister Chromatids- It is the identical copies of the DNA formed after the DNA replication i.e. in . The replicated copies are held together by a common centromere.
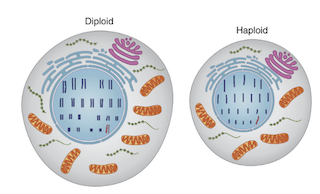
Haploid cell – The cell carrying a single set of chromosomes. Ex. haploid human cells like egg and sperm have 23 chromosomes.
Diploid cell – The cell carrying a double set of chromosomes. Ex. diploid human somatic cells have 23 + 23 chromosomes = 46.
Homologous chromosomes– In diploid cells, there are two sets of chromosomes. During sexual reproduction, the offspring receives one set of chromosome from both the parents (one is paternal and other is maternal). The chromosomes from both parental sets that have the same structure, size, gene loci and banding pattern are called homologous chromosomes.
Heterologous chromosomes – The chromosomes that do not have the same structure, size, gene loci and banding pattern as called heterologous chromosomes.
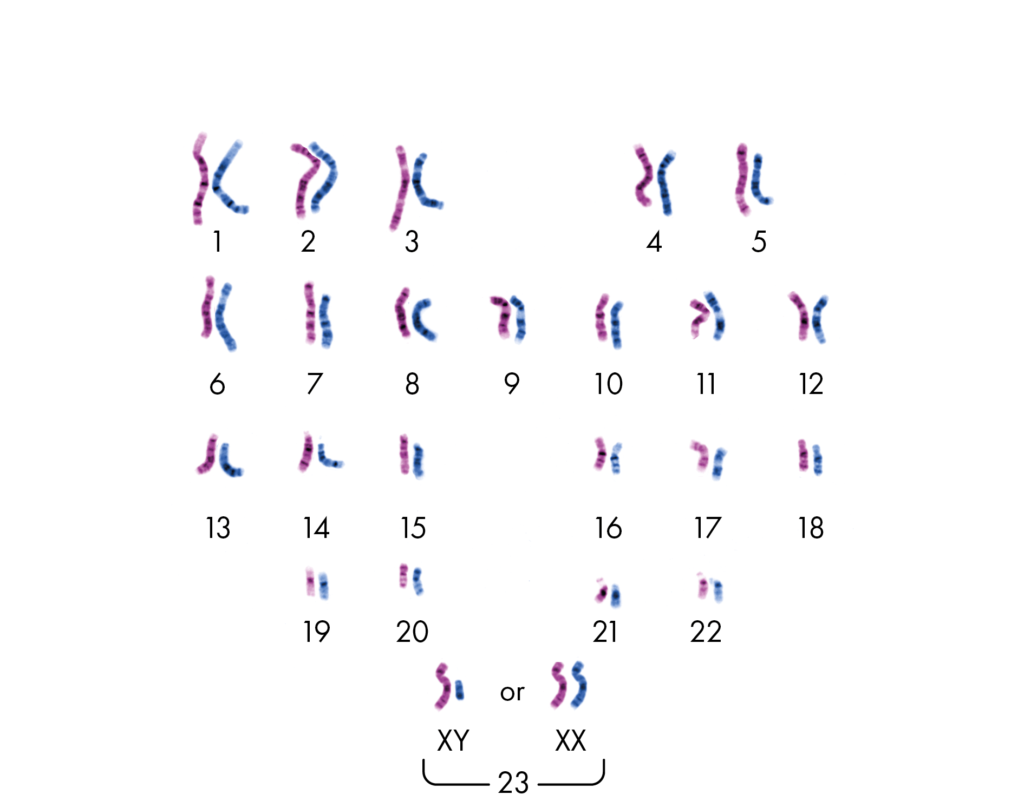
In the above image, the pink colored set of chromosomes is from Mom and the blue one is from dad. The chromosome with same structure, size, gene loci and banding pattern and paired together hence they are homologous chromosomes. The non paired chromosomes with different structure, size, gene loci and banding pattern are heterologous chromosomes.
Types of chromosomes –
All human normal chromosomes have two arms – p and q. ‘p’ comes from petit (french) which means small; ‘q’ name was chosen which alphabetically it is followed by p. The chromosomes have been divided based on the position of the constriction point i.e. centromere.
- Metacentric – The chromosome whose centromere is in middle, meaning p and q arms are of almost same length (e.g. chromosomes 1, 3, 16, 19, 20).
- Submetacentric – The chromosome whose centromere off-centre, which leads to shorter p arm as compared to q arm (e.g. chromosomes 2, 4 – 12, 17, 18, X).
- Acrocentric – The chromosome whose centromere is towards the chromosome’s end, leading to much shorter p arm (e.g. chromosomes 13 – 15, 21, 22, Y).
- Telocentric – The chromosome whose centromere is found at the end of the chromosome. Hence, there is no p arm (The telocentric chromosome are not found in humans).
Packaging of Chromosomes –
The diploid human cell contains 23 pairs of chromosomes which contains approximately 6 billion bp of DNA per cell. According to the Watson model of DNA, the base pair is 0.34nm long. Hence, one single human cell contains 2m long DNA [(0.34 × 10-9) × (6 × 109)]. Can you Imagine how this 2m DNA is present in a microscopic cell? And how the DNA is made available for gene expression. Moreover, we have around 50 trillion cells. It means the length of the total DNA would be 100 trillion meters. This length is twice the diameter of the solar system. This is possible only because of condensation or compaction of DNA. It is similar to the weaving thread wrapped around paper spool. While packaging the DNA, the linear DNA is condensed 1000 times than a linear DNA.
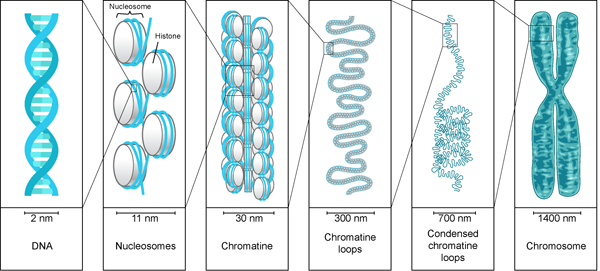
The DNA present in the nucleus is always found to be associated with proteins. This DNA-protein association is called chromatin. This is a less condensed form of DNA. It is found in the interphase of the Eukaryotic cell cycle, when the cell is not dividing. The chromatin structure allows the DNA replication and transcription.
Under electron microscope, the chromatin structure looks like beads on string, where the DNA is tightly associated with histone proteins. The DNA is wrapped around the core of histone proteins, and it appears like a bead. These tiny beads are called nucleosomes and their diameter is around 11nm.
While wrapping, 146 basepairs of the DNA show slightly less than two turns (1.7) around the histone core. Research has found that the AT region with minor groove are compressed while wrapping around the histone core. The AT regions allow easy bending of DNA Each histone core has two copies of H2A, H2B, H3 and H4 histone proteins hence forming histone octamer. The histone proteins are rich in positive amino acids and thus they allow negatively charged DNA to wrap around it. The DNA and histone proteins interact electrostatically and with non covalent bonds. When the nucleosomes are linked by an additional histone protein called H1 protein and linker DNA (20bp) it forms a chromatosome. Chromatosomes are the fundamental unit of chromatin structure. The core histone proteins’ N terminal tail extends out. The tails are subjected to covalent modification like acetylation, methylation or phosphorylation modelling and remodelling the chromatin structure.
The Formation of nucleosomes and chromosomes is the first stage of condensation. And this reduces the length of DNA by sevenfold i.e. one meter DNA becomes 14cm long. But this is still not sufficient to get accommodated in a microscopic cell. Hence, the chromatin needs to fold to more extent.
When chromatin is folded, packaged and condensed, it is called a chromosome. The structure of chromatin and chromosome is changeable and it is visible in different stages of the eukaryotic cell cycle.
The second level of compaction is ‘30 nm fiber’ because its diameter is 30nm. The beads on string folds in an irregular pattern forming 30nm fiber. The H1 protein plays a vital role in stabilizing 30 nm fiber. The 30nm fiber reduces the length of DNA by 50 times. The 30nm chromatin fiber behaves like a fluid mosaic showing different variations of the zig-zag model.This is possible due to varying length of linker DNA and H1 protein. In the zig zag model, the successive nucleosomes along with DNA are stacked alternatively. Such close proximity of nucleosomes favours interaction of adjacent nucleosomes’s histone protein tail.
The 30nm fiber is further folded to thick fibers forming 300nm to 700nm condensed chromatin loop and finally to 1400nm chromosome. This is achieved by forming chromatin loops around the scaffold protein. Such compaction, condenses the DNA from 1cm to 1um i.e. the packing ratio is 10,000:1.

The compaction of DNA hampers the molecular process like replication and transcription. In order to process the molecular processes, the DNA needs to unwind from the histone octamer (nucleosome) and open the helix and expose the template to the enzymes. The unwinding of DNA is achieved by modification of amino acid groups like acetylation, methylation and phosphorylation of histone protein N terminal tails. The modification of tails destabilises the non-covalent bonds between DNA and histone proteins allowing the unwrapping of DNA. This is called modelling of chromatin. This process is reversible and allows remodelling of chromatin. The modelling and remodelling is an energy driven process.
The packaging of DNA not only allows DNA to be accommodated in a small space but also permits gene expression. While looping, the related functional genes are brought together in close proximity to fulfil the need of the cell. While DNA packaging, the highly condensed chromatin region is called Heterochromatin and less condensed chromatin region is called Euchromatin. In Heterochromatin, the gene expression is inhibited and active gene expression is observed in euchromatin.
References-
https://www.nature.com/scitable/topicpage/chromosomes-14121320/
https://www.nature.com/scitable/topicpage/dna-packaging-nucleosomes-and-chromatin-310/
Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Chromosomal DNA and Its Packaging in the Chromatin Fiber. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26834
Dr. Sangha Bijekar has 9 years of Teaching Experience at University level. She loves to get engage in teaching and learning process. She is into blogging from last two years. She intends to provide student friendly reading material. She is avid Dog Lover and animal rescuer. She is learned Bharatnatyam and Katthak Dancer. She is into biking and She also loves to cook.


One thought on “Fundamentals of chromosome and Packaging of DNA”