In central Dogma of life, we have studied that DNA replication, transcription and translation are vital molecular processes. In Eukaryotes, the transcription is followed by messenger RNA processing, where the pre-mRNA is processed to form mature m-RNA. The mature m-RNA not only become functional but also increases the half life time of m-RNA. In m-RNA processing, the pre-mRNA undergoes capping at 5’end, splicing and polyadenylation at 3’end.
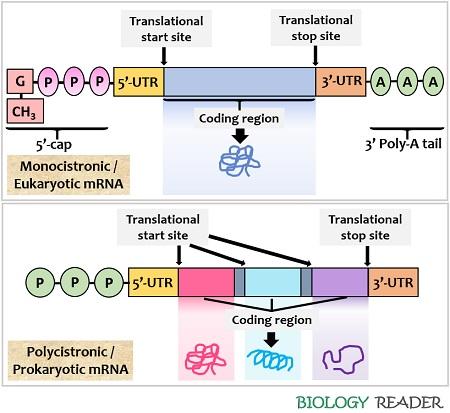
Let’s us compare the difference between the prokaryotic and eukaryotic m-RNA
Comparison between prokaryotic and Eukaryotic m-RNA –
| Prokaryotic m-RNA | Eukaryotic m-RNA |
| It is polycistronic producing several related functional proteins | It is monocistronic producing single protein |
| m-RNA is synthesized in cytoplasm | m-RNA is synthesized in nucleus |
| It neither have cap nor poly A tail | It contains both cap and poly tail |
| Transcription and Translation may occur simultaneously | Transcription occurs in nucleus followed by translation |
| Shine Dalgarno sequence of prokaryotic m-RNA is the ribosome binding site | 5’end cap of eukaryotic m-RNA is binding site for ribosome |
| Little or no modification or processing of prokaryotic mRNA | RNA processing like capping, splicing and polyadenylation occurs in eukaryotic m-RNA |
| Prokaryotic m-RNA has short life span of few seconds | Eukaryotic m-RNA can last for few hours. |
m-RNA Processing –
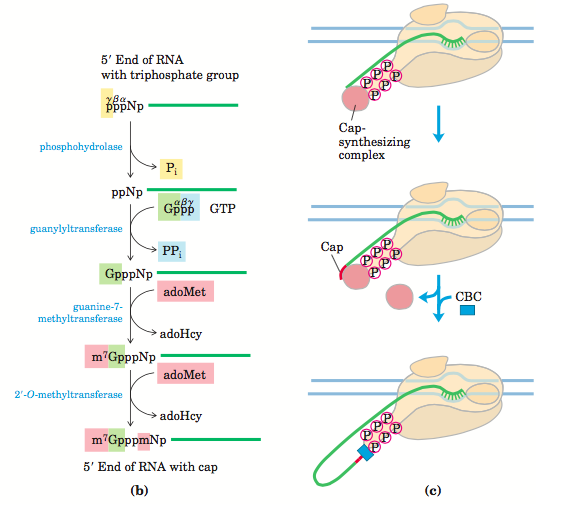
Capping of m-RNA-
The eukaryotic m-RNA is capped at 5’ end with 7 methylguanosine with 5’, 5’ triphosphate bond. The function of capping is to protect the m-RNA from Rnase. During translation, a specific protein called cap binding protein binds the cap of the m-RNA and plays a vital role in initiating the translation process. For adding the cap, the triphosphate of the last nucleotide at 5’end needs to be modified. For this, phosphohydrolase enzyme removes the gamma phosphate of the triphosphate and releasing PPi (two phosphorous). It is followed by the attachment of free guanine nucleotide to beta phosphate of last nucleotide at 5’end. This reaction is carried by guanyltransferase.
Addition of guanine is followed by methylation of N-7 of guanosine nucleotide. Additional methyl group is further added to the 2’OH group of the first and second nucleotides adjacent to the cap. The caping and elongation of transcription occurs simultaneously. This is achieved by collective and collaborative work of capping enzyme and CTD of RNA polymerase II.
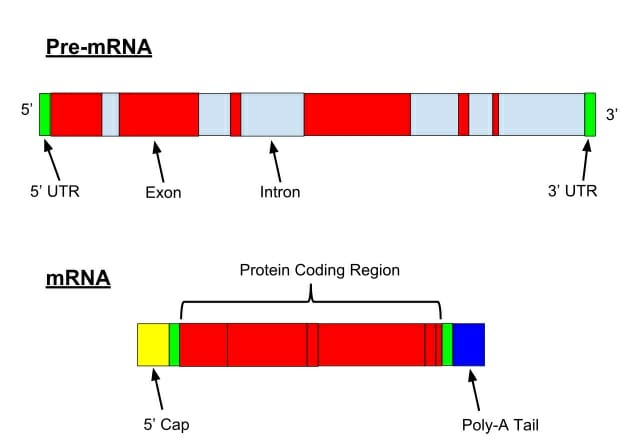
Splicing –
In eukaryotes, various RNA transcripts have introns (non-coding section) present in between exons (coding section). For m-RNA processing, the non-coding sections of pre-mRNA need to be removed before the translation process and it is called splicing. The splicing of introns and joining of exons of pre-mRNA produces mature m-RNA.
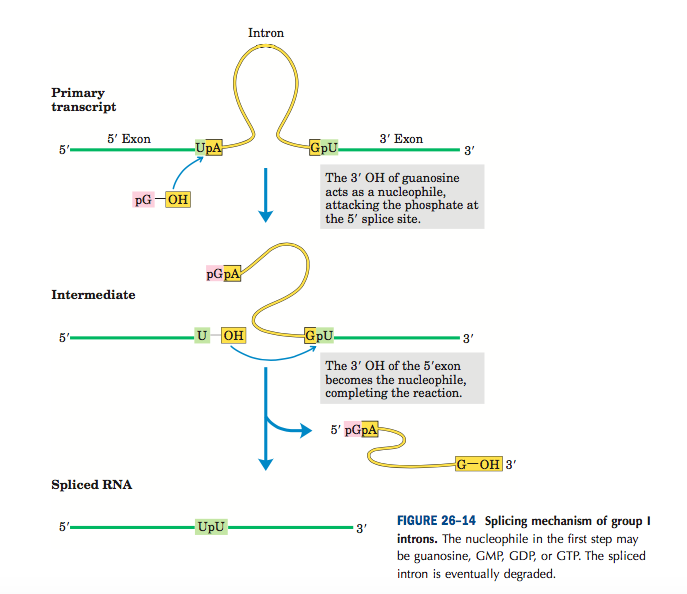
Group I intron splicing –
They are self-splicing types of introns where there is no need for any protein enzyme. Such group I introns are found in nuclear, mitochondrial and chloroplast RNA transcript. The group I intron splicing does not need any energy (ATP). The mechanism of self splicing is the nucleophilic attack of 2’ or 3’OH of one of the introns on the phosphate group of another end of the same intron. For group I splicing, guanine nucleotide is needed as a cofactor. The hydroxyl group of guanine nucleotide attacks on the 5’end splice site of the intron forming a 3′,5’ phosphodiester bond. The 3’OH of the exon attacks the 3′ splice site releasing the intron. The two exons are joined and form mature m-RNA.
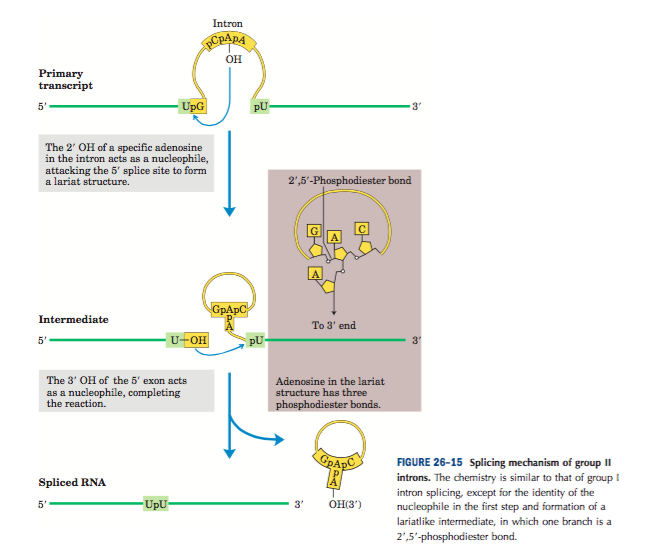
Group II Intron Splicing –
The group II introns are found in mitochondrial and chloroplast RNA transcript. The group II intron are of two types –
- Self-splicing
- Spliceosome
- Self Splicing – This group II introns can self spliced hence they do not need any enzyme or protein. They also do not need any energy for splicing. The mechanism of splicing is similar to the group I intron self splicing. In group II, adenine nucleotide is present in between the sequence of intron. The 2’OH group of adenine nucleotide attacks on the 5′ splice site of the intron forming lariat-like structure. This allows the 3’OH group of a 5’ splice site to attack on a phosphate group of 3’ splice site and release the intron. The two exons are joined and form mature m-RNA.
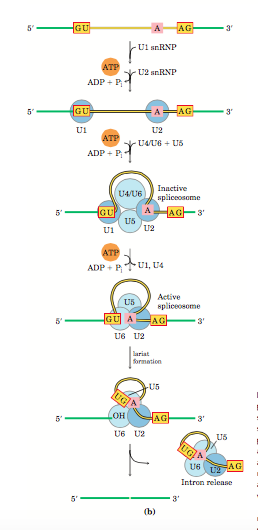
2. Spliceosome –
The spliceosome is a RNA and protein complex called as small nuclear ribonucleoproteins (SnRNPs pronounced as snurps). The RNA involved in SnRNPS are U1, U2, U4, U5 and U6, they are small and made up of 100 to 200 nucleotides. The SnRNPs catalyses and removes the introns from pre-mRNA.
SnRNP (snurps) = SnRNA (U1, U2, U4, U5, U6) +150 proteins.
SnRNP – Small nuclear ribonuclear proteins
SnRNA – Small nuclear RNA (100 t0 200 nucleotides long)
Dinucleotide GU at 5’end and AG at 3’ end of the intron acts like a landmark for splicing. The U1 SnRNP has a sequence complementary to GU, hence U1 binds it. U2 Auxiliary Proteins (U2AP) bind to the 5’ end of the intron and allow the binding of branch point binding (BPB) protein at the A residue (branch site). The Branch point binding protein is displaced by U2SnRNP and binds to A residue. The U2AP dissociates and U2 further recruits U4, U5 and U6 SnRNP forming an inactive spliceosome. Binding of U6SnRNP releases U1. U6 interacts with U2 and during this U4 is released forming an active spliceosome. The formation of a spliceosome requires energy in the form of ATP. The nucleophilic action of A residue forms the lariat structure. This is followed by the reaction between 5’ and 3’ splice site and complete the splicing reaction.
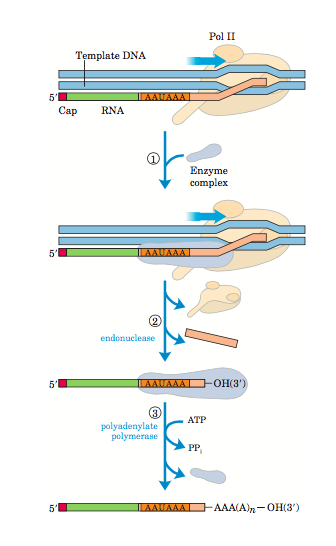
Addition of Poly A tail at 3’ end –
Most of the eukaryotic m-RNA have a poly A tail at its 3’end. It has been observed that a poly A tail has binding sites for many specific proteins that may assist in initiating the translation process and for exporting the mature RNA. The role of poly A tail and its binding proteins is to protect the m-RNA from enzymatic destruction. In prokaryotes, the function of the poly A tail is reverse, where it stimulates the m-RNA degradation. The addition of a poly A tail is a multistep process. While transcribing, the RNA polymerase II transcribes the RNA beyond the cleavage signal sequences. The cleavage site has two conservative sequences – 5’AAUAAA3’ (10 to 30 nucleotides at the upstream side of cleavage site) and GU residues rich (20 to 40 nucleotides athe the downstream side of cleavage site). The polyadenylation of m-RNA is catalysed by the protein complex with endonuclease and poly A polymerase activity. The pre-mRNA is cleaved at the cleavage site by endonuclease activity of the protein complex. The cleavage releases the functional pre-mRNA with free 3’OH group but still remains attached to RNA polymerase. The poly A polymerase (PAP) activity of the same protein complex adds around 200 Adenine residue at 3’OH forming a poly A tail.
References –
https://www.ncbi.nlm.nih.gov/books/NBK26850/
Principle of Biochemistry, Nelson and Cox, Lehninger, 5th Edition
Dr. Sangha Bijekar has 9 years of Teaching Experience at University level. She loves to get engage in teaching and learning process. She is into blogging from last two years. She intends to provide student friendly reading material. She is avid Dog Lover and animal rescuer. She is learned Bharatnatyam and Katthak Dancer. She is into biking and She also loves to cook.

